RECOMB proceedings will be accessible to registered participants:
Free access to RECOMB proceedings, courtesy of Springer until May 19th
Program timetable
Saturday April 21st
RECOMB I
-
Registration
-
-
Welcome Address
-
-
Peter Campbell (Keynote)
-
Clonal dynamics of normal human blood production
-
1. Cancer
-
Chair: Ben Raphael
-
Coffee break
-
-
2. Sequencing
-
Chair: Veli Makinen
-
Lunch break
-
-
Nevan Krogan (Keynote)
-
-
3. Proteomics session
-
Chair: Niko Beerenwinkel
-
Coffee break
-
-
4. Deep Learning
-
Chair: Jian Ma
Sponsored by IBM Research
Sunday April 22rd
RECOMB II
-
Ron Shamir (Keynote)
-
Disease bioinformatics: the good, the bad and the ugly
-
5. Cross-Species Functional Genomics
-
Chair: Cenk Sahinalp
-
Coffee break
-
-
6. Algorithmic foundations
-
Chair: Paul Medvedev
Sponsored by CNRS -
Lunch break
-
-
François Spitz (Keynote)
-
The genome in 3D: Function and regulation of long-distance relationships in development and disease
-
7. Genome Organization
-
Chair: Mathieu Blanchette
-
Coffee break
-
-
8. Cancer Phylogenetics
-
Chair: Layla Oesper
-
Poster Session
-
Odd numbers posters
-
Departure for social event
-
Monday April 23rd
RECOMB III
-
Tandy Warnow (Keynote)
-
Genome-scale estimation of the Tree of Life
-
9. Evolution
-
Chair: Teresa Przytycka
-
Coffee break
-
-
10. Genetics and Associations Studies
-
Chair: Russell Schwartz
-
Lunch break
-
-
Sarah Teichmann (Keynote)
-
Immunogenomics once cell at a time
-
11. Single-Cell Analysis
-
Chair: Bonnie Berger
-
Coffee break
-
-
12. Metagenomics & Microbiome
-
Chair: Fereydoun Hormozdiari
-
Poster Session
-
Even numbers posters
- Business meeting
-
Tuesday April 24th
RECOMB IV
-
13. Functional Genomics and Metagenomics
-
Chair: Alex Schoenhuth
-
Coffee break
-
-
14. Learning and Inference
-
Chair: Fabio Vandin
-
Lunch break
-
-
15. RNA
-
Chair: Mireille Regnier
-
Awards Ceremony and Closing Remarks
-
The scientific program also includes 150 selected posters.
Session 1 – Cancer
-
Chair: Ben Raphael
- ModulOmics: Integrating Multi-Omics Data to Identify Cancer Driver Modules
- Efficient Algorithms to Discover Alterations with Complementary Functional Association in Cancer
Session 2 – Sequencing
-
Chair: Veli Makinen
- Latent variable model for aligning barcoded short-reads improves downstream analyses
- Targeted Genotyping of Variable Number Tandem Repeats with adVNTR
- Assembly of Long Error-Prone Reads and Repeat Classification
- Mantis: A Fast, Small, and Exact Large-Scale Sequence-Search Index
Session 3 – Proteomics
-
Chair: Niko Beerenwinkel
- Constrained De Novo Sequencing of neo-Epitope Peptides using Tandem Mass Spectrometry
- [Highlight] Increased diversity of peptidic natural products revealed by modification-tolerant database search of mass spectra
Session 4 – Deep learning – Sponsored by IBM Research
-
Chair: Jian Ma
- Deciphering signaling specificity with deep neural networks
- Positive-unlabeled convolutional neural networks for particle picking in cryo-electron micrographs
- Deep learning reveals many more inter-protein residue-residue contacts than direct coupling analysis
- [Highlight] De novo Peptide Sequencing by Deep Learning
Session 5 – Cross-species Functional Genomics
-
Chair: Cenk Sahinalp
- A Multi-Species Functional Embedding Integrating Sequence and Network Structure
- Continuous-trait probabilistic model for comparing multi-species functional genomic data
Session 6 – Algorithmic foundations – Sponsored by CNRS
-
Chair: Paul Medvedev
- Algorithmic Framework for Approximate Matching Under Bounded Edits with Applications to Sequence Analysis
- Using Minimum Path Cover to Boost Dynamic Programming on DAGs: Co-Linear Chaining Extended
- GTED: Graph Traversal Edit Distance
- Reverse de Bruijn: Utilizing Reverse Peptide Synthesis to Cover All Amino Acid k-mers
Session 7 – Genome Organization
-
Chair: Mathieu Blanchette
- Contribution of structural variation to genome structure: TAD fusion discovery and ranking
- [Highlight] Tracking the evolution of 3D gene organization demonstrates its connection to phenotypic divergence
Session 8 – Cancer Phylogenetics
-
Chair: Layla Oesper
- SCIPhI: Single-cell mutation identification via phylogenetic inference
- Integrative inference of subclonal tumor evolution from single-cell and bulk sequencing data
Session 9 – Evolution
-
Chair: Teresa Przytycka
- Circular Networks from Distorted Metrics
- Modeling Dependence in Evolutionary Inference for Proteins
Session 10 – Genetics and Association Studies
-
Chair: Russell Schwartz
- Inference of population structure from ancient DNA
- A unifying framework for summary statistics imputation
- Tensor Composition Analysis Detects Cell-Type Specific Associations in Epigenetic Studies
- [Highlight] Prioritizing Tests of Epistasis Through Hierarchical Representation of Genomic Redundancies
Session 11 – Single-cell Analysis
-
Chair: Bonnie Berger
- Generalizable visualization of mega-scale single-cell data
- Probabilistic Count Matrix Factorization for Single Cell Expression Data Analysis
Session 12 – Metagenomics and Microbiome
-
Chair: Fereydoun Hormozdiari
- Long Reads Enable Accurate Estimates Of Complexity Of Metagenomes
- Accurate Reconstruction of Microbial Strains Using Representative Reference Genomes
- [Highlight] De Novo Viral Quasispecies Assembly using Overlap Graphs
Session 13 – Functional Genomics and Metagenomics
-
Chair: Alex Schoenhuth
- Assembly-free and alignment-free barcoding from genome skims
- Characterizing protein-DNA binding event subtypes in ChIP-exo data
- Chromatyping: Reconstructing nucleosome profiles from NOMe sequencing data
- [Highlight] Quantitating translational control: mRNA abundance-dependent and independent contributions and the mRNA sequences that specify them
Session 14 – Learning and Inference
-
Chair: Fabio Vandin
- Context-Specific Nested Effects Models
- Statistical inference of peroxisome dynamics
- Loss-function learning for digital tissue deconvolution
- [Highlight] An Immune Clock of Human Pregnancy
Session 15 – RNA
-
Chair: Mireille Regnier
- Fixed-Parameter Tractable Sampling for RNA Design with Multiple Target Structures
- AptaBlocks: Accelerating the Design of RNA-based Drug Delivery Systems
- A nested 2-level cross-validation ensemble learning pipeline suggests a negative pressure against crosstalk snoRNA-mRNA interactions in Saccharomyces Cerevisae
- Designing RNA Secondary Structures is Hard
Social event
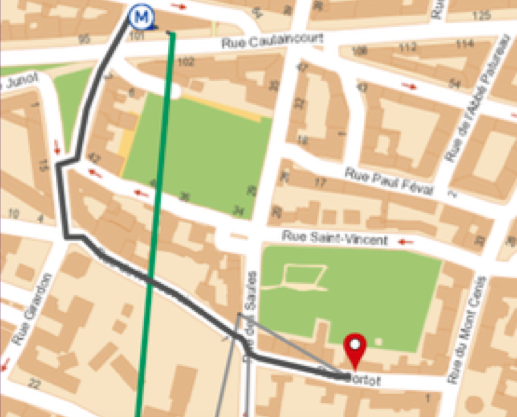
Our social event will be held at Musée Montmartre and gardens of Renoir, a meeting place for many artists including Auguste Renoir and Suzanne Paladin.
Starting from 7 PM, we will enjoy the Van Dongen Exhibition, a cocktail for all tastes, including a wine tasting accompanied by smoked salmon appetizers starting.
To get there, take the metro line 10 at Jussieu towards Boulogne/Pont de Saint-Cloud, and get off at Sèvres-Babylone. Switch to line 12 towards Front Populaire, and get off at Lamarck-Caulaincourt. Then walk 7 minutes un-til the 12 rue Cortot. Remember to buy your train tickets in advance to avoid queuing at the ticket machine with around 400 other scientists!